Xarray maps¶
In this example, we will start exporting eReefs data using the Xarray library and do similar visualisation as the ones we did with Numpy.
Load the required Python libraries¶
First of all, load the necessary libraries. These are the ones we discussed previously:
numpy
matplotlib
cartopy
xarray
import os
import numpy as np
import xarray as xr
import cartopy
import cartopy.crs as ccrs
import cartopy.feature as cfeature
from cartopy.mpl.gridliner import LONGITUDE_FORMATTER, LATITUDE_FORMATTER
cartopy.config['data_dir'] = os.getenv('CARTOPY_DIR', cartopy.config.get('data_dir'))
import cmocean
from matplotlib import pyplot as plt
#%config InlineBackend.figure_format = 'retina'
plt.ion() # To trigger the interactive inline mode
import warnings
warnings.filterwarnings("ignore", category=RuntimeWarning)
Build multi-file dataset¶
We will use the open_mfdataset function from xArray to open multiple netCDF files into a single xarray Dataset.
We will query load the GBR4km dataset from the AIMS server, so let’s first define the base URL:
# For the bio dataset
base_url = "http://thredds.ereefs.aims.gov.au/thredds/dodsC/s3://aims-ereefs-public-prod/derived/ncaggregate/ereefs/GBR4_H2p0_B3p1_Cq3b_Dhnd/daily-monthly/EREEFS_AIMS-CSIRO_GBR4_H2p0_B3p1_Cq3b_Dhnd_bgc_daily-monthly-"
# For the hydro dataset
base_url2 = "http://thredds.ereefs.aims.gov.au/thredds/dodsC/s3://aims-ereefs-public-prod/derived/ncaggregate/ereefs/gbr4_v2/daily-monthly/EREEFS_AIMS-CSIRO_gbr4_v2_hydro_daily-monthly-"
For the sake of the demonstration, we will only use 1 month:
month_st = 1 # Starting month
month_ed = 1 # Ending month
year = 2018 # Year
# Based on the server the file naming convention
biofiles = [f"{base_url}{year}-{month:02}.nc" for month in range(month_st, month_ed+1)]
hydrofiles = [f"{base_url2}{year}-{month:02}.nc" for month in range(month_st, month_ed+1)]
Loading dataset into xArray¶
Using xArray, we open these files into a Dataset:
ds_bio = xr.open_mfdataset(biofiles)
ds_hydro = xr.open_mfdataset(hydrofiles)
ds_bio
<xarray.Dataset>
Dimensions: (k: 17, latitude: 723, longitude: 491, time: 31)
Coordinates:
zc (k) float64 dask.array<chunksize=(17,), meta=np.ndarray>
* time (time) datetime64[ns] 2018-01-01T02:00:00 ... 2018-01-31...
* latitude (latitude) float64 -28.7 -28.67 -28.64 ... -7.066 -7.036
* longitude (longitude) float64 142.2 142.2 142.2 ... 156.8 156.8 156.9
Dimensions without coordinates: k
Data variables: (12/101)
alk (time, k, latitude, longitude) float32 dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
BOD (time, k, latitude, longitude) float32 dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
Chl_a_sum (time, k, latitude, longitude) float32 dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
CO32 (time, k, latitude, longitude) float32 dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
DIC (time, k, latitude, longitude) float32 dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
DIN (time, k, latitude, longitude) float32 dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
... ...
SGH_N (time, latitude, longitude) float32 dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
SGH_N_pr (time, latitude, longitude) float32 dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
SGHROOT_N (time, latitude, longitude) float32 dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
SGROOT_N (time, latitude, longitude) float32 dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
TSSM (time, latitude, longitude) float32 dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
Zenith2D (time, latitude, longitude) float32 dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
Attributes: (12/20)
Conventions: CF-1.0
NCO: netCDF Operators version 4.7.7 (Homepage...
RunID: 2
_CoordSysBuilder: ucar.nc2.dataset.conv.CF1Convention
aims_ncaggregate_buildDate: 2020-08-21T23:07:30+10:00
aims_ncaggregate_datasetId: products__ncaggregate__ereefs__GBR4_H2p0...
... ...
paramfile: /home/bai155/EMS_solar2/gbr4_H2p0_B3p1_C...
paramhead: eReefs 4 km grid. SOURCE Catchments with...
technical_guide_link: https://eatlas.org.au/pydio/public/aims-...
technical_guide_publish_date: 2020-08-18
title: eReefs AIMS-CSIRO GBR4 BioGeoChemical 3....
DODS_EXTRA.Unlimited_Dimension: time- k: 17
- latitude: 723
- longitude: 491
- time: 31
- zc(k)float64dask.array<chunksize=(17,), meta=np.ndarray>
- long_name :
- Z coordinate
- _CoordinateAxisType :
- Height
- _CoordinateZisPositive :
- up
- units :
- m
- positive :
- up
- axis :
- Z
- coordinate_type :
- Z
Array Chunk Bytes 136 B 136 B Shape (17,) (17,) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - time(time)datetime64[ns]2018-01-01T02:00:00 ... 2018-01-...
- long_name :
- Time
- standard_name :
- time
- coordinate_type :
- time
- _CoordinateAxisType :
- Time
- _ChunkSizes :
- 1024
array(['2018-01-01T02:00:00.000000000', '2018-01-02T02:00:00.000000000', '2018-01-03T02:00:00.000000000', '2018-01-04T02:00:00.000000000', '2018-01-05T02:00:00.000000000', '2018-01-06T02:00:00.000000000', '2018-01-07T02:00:00.000000000', '2018-01-08T02:00:00.000000000', '2018-01-09T02:00:00.000000000', '2018-01-10T02:00:00.000000000', '2018-01-11T02:00:00.000000000', '2018-01-12T02:00:00.000000000', '2018-01-13T02:00:00.000000000', '2018-01-14T02:00:00.000000000', '2018-01-15T02:00:00.000000000', '2018-01-16T02:00:00.000000000', '2018-01-17T02:00:00.000000000', '2018-01-18T02:00:00.000000000', '2018-01-19T02:00:00.000000000', '2018-01-20T02:00:00.000000000', '2018-01-21T02:00:00.000000000', '2018-01-22T02:00:00.000000000', '2018-01-23T02:00:00.000000000', '2018-01-24T02:00:00.000000000', '2018-01-25T02:00:00.000000000', '2018-01-26T02:00:00.000000000', '2018-01-27T02:00:00.000000000', '2018-01-28T02:00:00.000000000', '2018-01-29T02:00:00.000000000', '2018-01-30T02:00:00.000000000', '2018-01-31T02:00:00.000000000'], dtype='datetime64[ns]') - latitude(latitude)float64-28.7 -28.67 ... -7.066 -7.036
- units :
- degrees_north
- long_name :
- Latitude
- standard_name :
- latitude
- projection :
- geographic
- coordinate_type :
- latitude
- _CoordinateAxisType :
- Lat
array([-28.696022, -28.666022, -28.636022, ..., -7.096022, -7.066022, -7.036022]) - longitude(longitude)float64142.2 142.2 142.2 ... 156.8 156.9
- units :
- degrees_east
- long_name :
- Longitude
- standard_name :
- longitude
- projection :
- geographic
- coordinate_type :
- longitude
- _CoordinateAxisType :
- Lon
array([142.168788, 142.198788, 142.228788, ..., 156.808788, 156.838788, 156.868788])
- alk(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- alk
- units :
- mmol m-3
- long_name :
- Total alkalinity
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - BOD(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- BOD
- units :
- mg O m-3
- long_name :
- Biochemical Oxygen Demand
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - Chl_a_sum(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- Chl_a_sum
- units :
- mg Chl m-3
- long_name :
- Total Chlorophyll
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - CO32(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- CO32
- units :
- mmol m-3
- long_name :
- Carbonate
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - DIC(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- DIC
- units :
- mg C m-3
- long_name :
- Dissolved Inorganic Carbon
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - DIN(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- DIN
- units :
- mg N m-3
- long_name :
- Dissolved Inorganic Nitrogen
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - DIP(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- DIP
- units :
- mg P m-3
- long_name :
- Dissolved Inorganic Phosphorus
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - DOR_C(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- DOR_C
- units :
- mg C m-3
- long_name :
- Dissolved Organic Carbon
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - DOR_N(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- DOR_N
- units :
- mg N m-3
- long_name :
- Dissolved Organic Nitrogen
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - DOR_P(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- DOR_P
- units :
- mg P m-3
- long_name :
- Dissolved Organic Phosphorus
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - Dust(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- Dust
- units :
- kg m-3
- long_name :
- Dust
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - EFI(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- EFI
- units :
- kg m-3
- long_name :
- Ecology Fine Inorganics
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - FineSed(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- FineSed
- units :
- kg m-3
- long_name :
- FineSed
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - Fluorescence(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- Fluorescence
- units :
- mg chla m-3
- long_name :
- Simulated Fluorescence
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - HCO3(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- HCO3
- units :
- mmol m-3
- long_name :
- Bicarbonate
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - Kd_490(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- Kd_490
- units :
- m-1
- long_name :
- Vert. att. at 490 nm
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - MPB_Chl(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- MPB_Chl
- units :
- mg Chl m-3
- long_name :
- Microphytobenthos chlorophyll
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - MPB_N(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- MPB_N
- units :
- mg N m-3
- long_name :
- Microphytobenthos N
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - Mud-carbonate(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- Mud-carbonate
- units :
- kg m-3
- long_name :
- Mud-carbonate
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - Mud-mineral(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- Mud-mineral
- units :
- kg m-3
- long_name :
- Mud-mineral
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - Nfix(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- Nfix
- units :
- mg N m-3 s-1
- long_name :
- N2 fixation
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - NH4(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- NH4
- units :
- mg N m-3
- long_name :
- Ammonia
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - NO3(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- NO3
- units :
- mg N m-3
- long_name :
- Nitrate
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - omega_ar(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- omega_ar
- units :
- nil
- long_name :
- Aragonite saturation state
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - Oxy_sat(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- Oxy_sat
- units :
- %
- long_name :
- Oxygen saturation percent
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - Oxygen(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- Oxygen
- units :
- mg O m-3
- long_name :
- Dissolved Oxygen
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - P_Prod(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- P_Prod
- units :
- mg C m-3 d-1
- long_name :
- Phytoplankton total productivity
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - PAR(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- PAR
- units :
- mol photon m-2 s-1
- long_name :
- Av. PAR in layer
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - PAR_z(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- PAR_z
- units :
- mol photon m-2 s-1
- long_name :
- Downwelling PAR at top of layer
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - pco2surf(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- pco2surf
- units :
- ppmv
- long_name :
- oceanic pCO2 (ppmv)
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - PH(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- PH
- units :
- log(mM)
- long_name :
- PH
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - PhyL_Chl(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- PhyL_Chl
- units :
- mg Chl m-3
- long_name :
- Large Phytoplankton chlorophyll
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - PhyL_N(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- PhyL_N
- units :
- mg N m-3
- long_name :
- Large Phytoplankton N
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - PhyS_Chl(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- PhyS_Chl
- units :
- mg Chl m-3
- long_name :
- Small Phytoplankton chlorophyll
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - PhyS_N(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- PhyS_N
- units :
- mg N m-3
- long_name :
- Small Phytoplankton N
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - PhyS_NR(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- PhyS_NR
- units :
- mg N m-3
- long_name :
- Small Phytoplankton N reserve
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - PIP(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- PIP
- units :
- mg P m-3
- long_name :
- Particulate Inorganic Phosphorus
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - salt(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- salt
- units :
- PSU
- long_name :
- Salinity
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - TC(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- TC
- units :
- mg C m-3
- long_name :
- Total C
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - temp(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- temp
- units :
- degrees C
- long_name :
- Temperature
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - TN(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- TN
- units :
- mg N m-3
- long_name :
- Total N
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - TP(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- TP
- units :
- mg P m-3
- long_name :
- Total P
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - Tricho_Chl(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- Tricho_Chl
- units :
- mg Chl m-3
- long_name :
- Trichodesmium chlorophyll
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - Tricho_N(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- Tricho_N
- units :
- mg N m-3
- long_name :
- Trichodesmium Nitrogen
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - Z_grazing(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- Z_grazing
- units :
- mg C m-3 d-1
- long_name :
- Zooplankton total grazing
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - ZooL_N(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- ZooL_N
- units :
- mg N m-3
- long_name :
- Large Zooplankton N
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - ZooS_N(time, k, latitude, longitude)float32dask.array<chunksize=(31, 17, 723, 491), meta=np.ndarray>
- short_name :
- ZooS_N
- units :
- mg N m-3
- long_name :
- Small Zooplankton N
- _ChunkSizes :
- [ 1 1 133 491]
Array Chunk Bytes 748.33 MB 748.33 MB Shape (31, 17, 723, 491) (31, 17, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - CH_N(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- CH_N
- units :
- g N m-2
- long_name :
- Coral host N
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - CS_bleach(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- CS_bleach
- units :
- d-1
- long_name :
- Coral bleach rate
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - CS_Chl(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- CS_Chl
- units :
- mg Chl m-2
- long_name :
- Coral symbiont Chl
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - CS_N(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- CS_N
- units :
- mg N m-2
- long_name :
- Coral symbiont N
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - EpiPAR_sg(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- EpiPAR_sg
- units :
- mol photon m-2 d-1
- long_name :
- Light intensity above seagrass
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - eta(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- eta
- units :
- metre
- long_name :
- Surface Elevation
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - MA_N(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- MA_N
- units :
- g N m-2
- long_name :
- Macroalgae N
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - MA_N_pr(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- MA_N_pr
- units :
- mg N m-2 d-1
- long_name :
- Macroalgae net production
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - month_EpiPAR_sg(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- month_EpiPAR_sg
- units :
- mol photon m-2
- long_name :
- Monthly dose light above seagrass
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - R_400(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- R_400
- units :
- sr-1
- long_name :
- Remote-sensing reflectance @ 400 nm
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - R_410(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- R_410
- units :
- sr-1
- long_name :
- Remote-sensing reflectance @ 410 nm
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - R_412(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- R_412
- units :
- sr-1
- long_name :
- Remote-sensing reflectance @ 412 nm
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - R_443(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- R_443
- units :
- sr-1
- long_name :
- Remote-sensing reflectance @ 443 nm
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - R_470(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- R_470
- units :
- sr-1
- long_name :
- Remote-sensing reflectance @ 470 nm
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - R_486(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- R_486
- units :
- sr-1
- long_name :
- Remote-sensing reflectance @ 486 nm
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - R_488(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- R_488
- units :
- sr-1
- long_name :
- Remote-sensing reflectance @ 488 nm
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - R_490(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- R_490
- units :
- sr-1
- long_name :
- Remote-sensing reflectance @ 490 nm
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - R_510(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- R_510
- units :
- sr-1
- long_name :
- Remote-sensing reflectance @ 510 nm
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - R_531(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- R_531
- units :
- sr-1
- long_name :
- Remote-sensing reflectance @ 531 nm
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - R_547(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- R_547
- units :
- sr-1
- long_name :
- Remote-sensing reflectance @ 547 nm
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - R_551(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- R_551
- units :
- sr-1
- long_name :
- Remote-sensing reflectance @ 551 nm
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - R_555(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- R_555
- units :
- sr-1
- long_name :
- Remote-sensing reflectance @ 555 nm
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - R_560(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- R_560
- units :
- sr-1
- long_name :
- Remote-sensing reflectance @ 560 nm
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - R_590(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- R_590
- units :
- sr-1
- long_name :
- Remote-sensing reflectance @ 590 nm
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - R_620(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- R_620
- units :
- sr-1
- long_name :
- Remote-sensing reflectance @ 620 nm
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - R_640(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- R_640
- units :
- sr-1
- long_name :
- Remote-sensing reflectance @ 640 nm
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - R_645(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- R_645
- units :
- sr-1
- long_name :
- Remote-sensing reflectance @ 645 nm
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - R_665(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- R_665
- units :
- sr-1
- long_name :
- Remote-sensing reflectance @ 665 nm
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - R_667(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- R_667
- units :
- sr-1
- long_name :
- Remote-sensing reflectance @ 667 nm
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - R_671(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- R_671
- units :
- sr-1
- long_name :
- Remote-sensing reflectance @ 671 nm
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - R_673(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- R_673
- units :
- sr-1
- long_name :
- Remote-sensing reflectance @ 673 nm
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - R_678(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- R_678
- units :
- sr-1
- long_name :
- Remote-sensing reflectance @ 678 nm
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - R_681(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- R_681
- units :
- sr-1
- long_name :
- Remote-sensing reflectance @ 681 nm
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - R_709(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- R_709
- units :
- sr-1
- long_name :
- Remote-sensing reflectance @ 709 nm
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - R_745(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- R_745
- units :
- sr-1
- long_name :
- Remote-sensing reflectance @ 745 nm
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - R_748(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- R_748
- units :
- sr-1
- long_name :
- Remote-sensing reflectance @ 748 nm
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - R_754(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- R_754
- units :
- sr-1
- long_name :
- Remote-sensing reflectance @ 754 nm
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - R_761(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- R_761
- units :
- sr-1
- long_name :
- Remote-sensing reflectance @ 761 nm
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - R_764(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- R_764
- units :
- sr-1
- long_name :
- Remote-sensing reflectance @ 764 nm
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - R_767(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- R_767
- units :
- sr-1
- long_name :
- Remote-sensing reflectance @ 767 nm
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - R_778(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- R_778
- units :
- sr-1
- long_name :
- Remote-sensing reflectance @ 778 nm
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - Secchi(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- Secchi
- units :
- m
- long_name :
- Secchi from 488 nm
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - SG_N(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- SG_N
- units :
- g N m-2
- long_name :
- Seagrass N
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - SG_N_pr(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- SG_N_pr
- units :
- mg N m-2 d-1
- long_name :
- Seagrass net production
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - SG_shear_mort(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- SG_shear_mort
- units :
- g N m-2 d-1
- long_name :
- Seagrass shear stress mort
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - SGD_N(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- SGD_N
- units :
- g N m-2
- long_name :
- Deep seagrass N
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - SGD_N_pr(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- SGD_N_pr
- units :
- mg N m-2 d-1
- long_name :
- Deep seagrass net production
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - SGD_shear_mort(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- SGD_shear_mort
- units :
- g N m-2 d-1
- long_name :
- Deep seagrass shear stress mort
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - SGH_N(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- SGH_N
- units :
- g N m-2
- long_name :
- Halophila N
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - SGH_N_pr(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- SGH_N_pr
- units :
- mg N m-2 d-1
- long_name :
- Halophila net production
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - SGHROOT_N(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- SGHROOT_N
- units :
- g N m-2
- long_name :
- Halophila root N
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - SGROOT_N(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- SGROOT_N
- units :
- g N m-2
- long_name :
- Seagrass root N
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - TSSM(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- TSSM
- units :
- g TSS m-3
- long_name :
- TSS from 645 nm (Petus et al., 2014)
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray - Zenith2D(time, latitude, longitude)float32dask.array<chunksize=(31, 723, 491), meta=np.ndarray>
- short_name :
- Zenith2D
- units :
- rad
- long_name :
- Solar zenith
- _ChunkSizes :
- [ 1 133 491]
Array Chunk Bytes 44.02 MB 44.02 MB Shape (31, 723, 491) (31, 723, 491) Count 2 Tasks 1 Chunks Type float32 numpy.ndarray
- Conventions :
- CF-1.0
- NCO :
- netCDF Operators version 4.7.7 (Homepage = http://nco.sf.net, Code = http://github.com/nco/nco)
- RunID :
- 2
- _CoordSysBuilder :
- ucar.nc2.dataset.conv.CF1Convention
- aims_ncaggregate_buildDate :
- 2020-08-21T23:07:30+10:00
- aims_ncaggregate_datasetId :
- products__ncaggregate__ereefs__GBR4_H2p0_B3p1_Cq3b_Dhnd__daily-monthly/EREEFS_AIMS-CSIRO_GBR4_H2p0_B3p1_Cq3b_Dhnd_bgc_daily-monthly-2018-01
- aims_ncaggregate_firstDate :
- 2018-01-01T12:00:00+10:00
- aims_ncaggregate_inputs :
- [products__ncaggregate__ereefs__GBR4_H2p0_B3p1_Cq3b_Dhnd__raw/EREEFS_AIMS-CSIRO_GBR4_H2p0_B3p1_Cq3b_Dhnd_bgc_raw_2018-01::MD5:922cfd031369e604eab88561e411dc0e]
- aims_ncaggregate_lastDate :
- 2018-01-31T12:00:00+10:00
- codehead :
- CSIRO Environmental Modelling Suite
- description :
- Regridding of daily input data (from eReefs AIMS-CSIRO GBR4 BioGeoChemical 3.1 subset) from curvilinear (per input data) to rectilinear via inverse weighted distance from up to 4 closest cells.
- ems_version :
- v1.1.1 rev(6244M)
- history :
- Tue Oct 8 15:38:27 2019: ncatted -a positive,botz,o,char,up -a missing_value,botz,o,double,99. -a outside,botz,o,double,-9999. gbr4_bgc_all_simple_2018-01.nc 2020-08-20T23:45:30+10:00: vendor: AIMS; processing: None summaries 2020-08-21T23:07:30+10:00: vendor: AIMS; processing: None summaries
- metadata_link :
- https://eatlas.org.au/data/uuid/61f3a6df-2c4a-46b6-ab62-3f3a9bf4e87a
- paramfile :
- /home/bai155/EMS_solar2/gbr4_H2p0_B3p1_Cb/tran/GBR4_H2p0_B3p1_Cq3b_Dhnd.tran
- paramhead :
- eReefs 4 km grid. SOURCE Catchments with 2019 condition from Dec 1, 2010 to June,30, 2018, Empirical SOURCE with 2019 condition, Jul 1, 2018 to April 30, 2019. More details of naming protocol at: eReefs.info.
- technical_guide_link :
- https://eatlas.org.au/pydio/public/aims-ereefs-platform-technical-guide-to-derived-products-from-csiro-ereefs-models-pdf
- technical_guide_publish_date :
- 2020-08-18
- title :
- eReefs AIMS-CSIRO GBR4 BioGeoChemical 3.1 (baseline catchment conditions) daily aggregation
- DODS_EXTRA.Unlimited_Dimension :
- time
Davies Reef position:
reef_lat = -18.82
reef_lon = 147.64
Plotting Xarray dataset¶
We will use the Xarray plot function.
See also
Check out this link to see how to use Xarray’s convenient matplotlib-backed plotting interface to visualize your datasets!
# Figure size
size = (9, 10)
# Color from cmocean
color = cmocean.cm.balance
# Defining the figure
fig = plt.figure(figsize=size, facecolor='w', edgecolor='k')
# Axes with Cartopy projection
ax = plt.axes(projection=ccrs.PlateCarree())
# and extent
ax.set_extent([142.4, 157, -7, -28.6], ccrs.PlateCarree())
# Plotting using Matplotlib
# We plot the PH at the surface at the final recorded time interval
cf = ds_bio.PH.isel(time=-1,k=-1).plot(
transform=ccrs.PlateCarree(), cmap=color,
vmin = 7.975, vmax = 8.125,
add_colorbar=False
)
# Color bar
cbar = fig.colorbar(cf, ax=ax, fraction=0.027, pad=0.045,
orientation="horizontal")
cbar.set_label(ds_bio.PH.long_name+' '+ds_bio.PH.units, rotation=0,
labelpad=5, fontsize=10)
cbar.ax.tick_params(labelsize=8)
# Title
plt.title('zc = '+str(ds_bio.zc.values.item(-1))+' m at '+str(ds_bio.coords['time'].values[-1])[:10],
fontsize=11
)
# Plot lat/lon grid
gl = ax.gridlines(crs=ccrs.PlateCarree(), draw_labels=True,
linewidth=0.1, color='k', alpha=1,
linestyle='--')
gl.top_labels = False
gl.right_labels = False
gl.xformatter = LONGITUDE_FORMATTER
gl.yformatter = LATITUDE_FORMATTER
gl.xlabel_style = {'size': 8}
gl.ylabel_style = {'size': 8}
# Add map features with Cartopy
ax.add_feature(cfeature.NaturalEarthFeature('physical', 'land', '10m',
edgecolor='face',
facecolor='lightgray'))
ax.coastlines(linewidth=1)
# Site Davies Reef
ax.scatter(reef_lon, reef_lat, c='deeppink', s=50, edgecolors='k', linewidth=1, transform=ccrs.PlateCarree())
plt.tight_layout()
plt.show()
fig.clear()
plt.close(fig)
plt.clf()
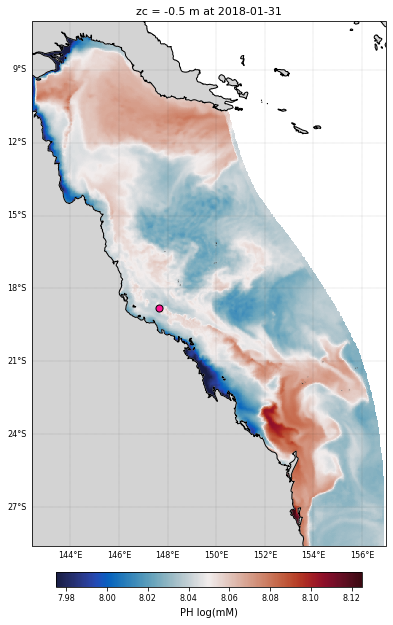
<Figure size 432x288 with 0 Axes>
Clip the Dataset¶
To reduce the Dataset size we will clip the spatial extent based on longitudinal and latitudinal values.
This is easely done using the sel function with the slice method.
min_lon = 146 # lower left longitude
min_lat = -21 # lower left latitude
max_lon = 150 # upper right longitude
max_lat = -16 # upper right latitude
# Defining the boundaries
lon_bnds = [min_lon, max_lon]
lat_bnds = [min_lat, max_lat]
# Performing the reduction
ds_bio_clip = ds_bio.sel(latitude=slice(*lat_bnds), longitude=slice(*lon_bnds))
ds_hydro_clip = ds_hydro.sel(latitude=slice(*lat_bnds), longitude=slice(*lon_bnds))
Let’s plot the clipped region using the same approach as above:
# Figure size
size = (9, 10)
# Color from cmocean
color = cmocean.cm.balance
# Defining the figure
fig = plt.figure(figsize=size, facecolor='w', edgecolor='k')
# Axes with Cartopy projection
ax = plt.axes(projection=ccrs.PlateCarree())
# and extent
ax.set_extent([min_lon, max_lon, min_lat, max_lat], ccrs.PlateCarree())
# Plotting using Matplotlib
# We plot the PH at the surface at the final recorded time interval
cf = ds_bio_clip.PH.isel(time=-1,k=-1).plot(
transform=ccrs.PlateCarree(), cmap=color,
vmin = 7.98, vmax = 8.08,
add_colorbar=False
)
# Color bar
cbar = fig.colorbar(cf, ax=ax, fraction=0.027, pad=0.045,
orientation="horizontal")
cbar.set_label(ds_bio_clip.PH.long_name+' '+ds_bio_clip.PH.units, rotation=0,
labelpad=5, fontsize=10)
cbar.ax.tick_params(labelsize=8)
# Title
plt.title('zc = '+str(ds_bio_clip.zc.values.item(-1))+' m at '+
str(ds_bio_clip.coords['time'].values[-1])[:10],
fontsize=11
)
# Plot lat/lon grid
gl = ax.gridlines(crs=ccrs.PlateCarree(), draw_labels=True,
linewidth=0.1, color='k', alpha=1,
linestyle='--')
gl.top_labels = False
gl.right_labels = False
gl.xformatter = LONGITUDE_FORMATTER
gl.yformatter = LATITUDE_FORMATTER
gl.xlabel_style = {'size': 8}
gl.ylabel_style = {'size': 8}
# Add map features with Cartopy
ax.add_feature(cfeature.NaturalEarthFeature('physical', 'land', '10m',
edgecolor='face',
facecolor='lightgray'))
ax.coastlines(linewidth=1)
# Site Davies Reef
ax.scatter(reef_lon, reef_lat, c='deeppink', s=70, edgecolors='k',
linewidth=1, transform=ccrs.PlateCarree())
plt.tight_layout()
plt.show()
fig.clear()
plt.close(fig)
plt.clf()
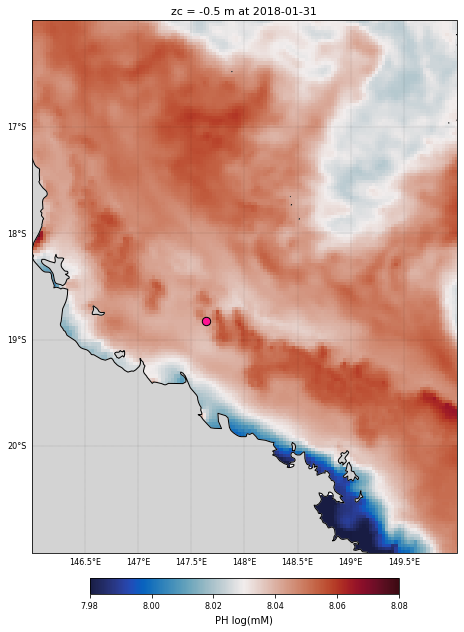
<Figure size 432x288 with 0 Axes>
Adding a quiver plot on top¶
To make the quiver plot, we will first resample the dataset and take one point every 7 times. It allows to have less arrows on the map making it more readable.
# Figure size
size = (7, 8)
# Time step to plot
timevar = 0
# z-coordinate position (here the top one)
zcvar = -1
# Color from cmocean
color = cmocean.cm.speed
# Defining the figure
fig = plt.figure(figsize=size, facecolor='w',
edgecolor='k')
# Axes with Cartopy projection
ax = plt.axes(projection=ccrs.PlateCarree())
# and extent
ax.set_extent([min_lon, max_lon, min_lat, max_lat],
ccrs.PlateCarree())
# Plotting using Matplotlib the mean current
cf = ds_hydro_clip.mean_cur.isel(time=timevar,k=zcvar).plot(
transform=ccrs.PlateCarree(), cmap=color,
vmin = 0.1, vmax = 0.7,
add_colorbar=False
)
# Resampling using the slice method
resample = ds_hydro_clip.isel(time=timevar,k=zcvar,longitude=slice(None, None, 7),
latitude=slice(None, None, 7))
# Defining the quiver plot
quiver = resample.plot.quiver(x='longitude', y='latitude', u='u', v='v',
transform=ccrs.PlateCarree(), scale=8)
# Vector options declaration
veclenght = 0.5
maxstr = '%3.1f m/s' % veclenght
plt.quiverkey(quiver,0.1,0.1,veclenght,maxstr,labelpos='S',
coordinates='axes').set_zorder(11)
# Color bar
cbar = fig.colorbar(cf, ax=ax, fraction=0.027, pad=0.045,
orientation="horizontal")
cbar.set_label(ds_hydro_clip.mean_cur.long_name+' '+
ds_hydro_clip.mean_cur.units, rotation=0,
labelpad=5, fontsize=10)
cbar.ax.tick_params(labelsize=8)
# Title
plt.title('zc = '+str(ds_hydro_clip.mean_cur.zc.values.item(zcvar))+' m at '+
str(ds_hydro_clip.mean_cur.coords['time'].values[timevar])[:10],
fontsize=11
)
# Plot lat/lon grid
gl = ax.gridlines(crs=ccrs.PlateCarree(), draw_labels=True,
linewidth=0.1, color='k', alpha=1,
linestyle='--')
gl.top_labels = False
gl.right_labels = False
gl.xformatter = LONGITUDE_FORMATTER
gl.yformatter = LATITUDE_FORMATTER
gl.xlabel_style = {'size': 8}
gl.ylabel_style = {'size': 8}
# Add map features with Cartopy
ax.add_feature(cfeature.NaturalEarthFeature('physical', 'land', '10m',
edgecolor='face',
facecolor='lightgray'))
ax.coastlines(linewidth=1)
# Site Davies Reef
ax.scatter(reef_lon, reef_lat, c='deeppink', s=50, edgecolors='k',
linewidth=1, transform=ccrs.PlateCarree()).set_zorder(11)
plt.tight_layout()
plt.show()
fig.clear()
plt.close(fig)
plt.clf()
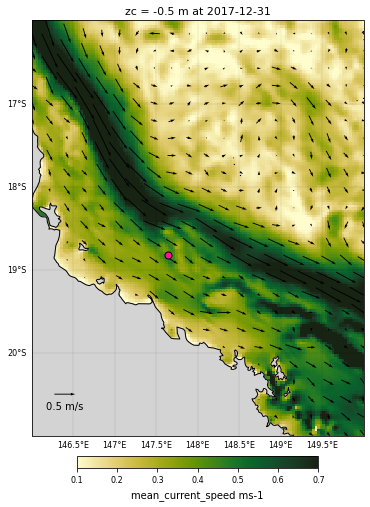
<Figure size 432x288 with 0 Axes>
Cross-sections on Xarray dataset¶
To define the cross-section, we chose to select (sel) several latitudes or longitudes and compute the mean along the opposite dimension:
# Section along the longitude
lonsec = ds_bio_clip.sel(latitude=slice(*[-19.,-18.5])).mean(dim=('latitude'),skipna=True)
# Section along the latitude
latsec = ds_bio_clip.sel(longitude=slice(*[147.2,148.])).mean(dim=('longitude'),skipna=True)
Let’s visualise the selected mean cross-sections:
# Figure size
size = (12, 4)
# Time step to plot
timevar = -1
# Color from cmocean
color = cmocean.cm.speed
# Defining the figure
fig = plt.figure(figsize=size, facecolor='w', edgecolor='k')
ax = plt.axes()
# Plotting using Matplotlib the temperature over the z-coordinate
sec = lonsec.temp.isel(time=timevar).plot(y="zc", add_colorbar=False)
# Color bar
cbar = fig.colorbar(sec, ax=ax, fraction=0.027, pad=0.045)
cbar.set_label(ds_bio.temp.long_name+' '+
ds_bio.temp.units, rotation=90,
labelpad=5, fontsize=10)
cbar.ax.tick_params(labelsize=8)
# Title
plt.title(str(lonsec.temp.coords['time'].values[timevar])[:10],
fontsize=11
)
# Site Davies Reef visualised as a line
ax.plot([reef_lon,reef_lon], [-1000,16], c='deeppink',
linewidth=3)
plt.tight_layout()
plt.show()
fig.clear()
plt.close(fig)
plt.clf()
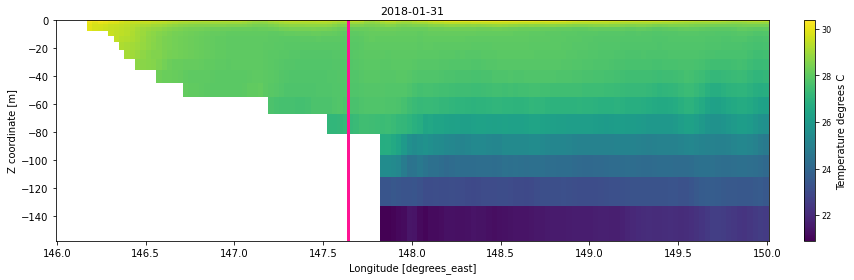
<Figure size 432x288 with 0 Axes>
# Figure size
size = (12, 4)
# Time step to plot
timevar = -1
# Color from cmocean
color = cmocean.cm.speed
# Defining the figure
fig = plt.figure(figsize=size, facecolor='w', edgecolor='k')
ax = plt.axes()
# Plotting using Matplotlib the temperature over the z-coordinate
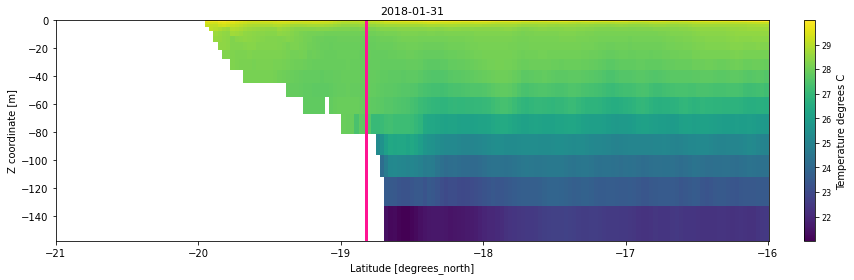
sec = latsec.temp.isel(time=timevar).plot(y="zc",add_colorbar=False)
# Color bar
cbar = fig.colorbar(sec, ax=ax, fraction=0.027, pad=0.045)
cbar.set_label(ds_bio.temp.long_name+' '+
ds_bio.temp.units, rotation=90,
labelpad=5, fontsize=10)
cbar.ax.tick_params(labelsize=8)
# Title
plt.title(str(latsec.temp.coords['time'].values[timevar])[:10],
fontsize=11
)
# Site Davies Reef visualised as a line
ax.plot([reef_lat,reef_lat], [-1000,16], c='deeppink',
linewidth=3)
plt.tight_layout()
plt.show()
fig.clear()
plt.close(fig)
plt.clf()
<Figure size 432x288 with 0 Axes>
