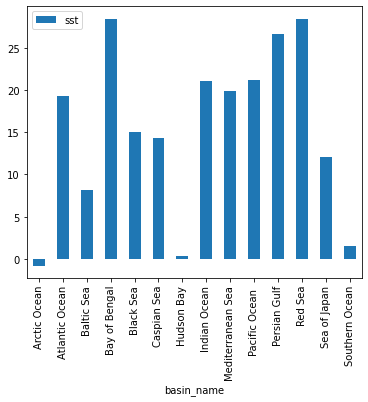
Advanced Xarray¶
This material is adapted from the Earth and Environmental Data Science, from Ryan Abernathey (Columbia University).
In this notebook, we cover some more advanced aspects of Xarray.
Groupby¶
Xarray copies Pandas’ very useful groupby functionality, enabling the “split / apply / combine” workflow on xarray DataArrays and Datasets. In the first part of the notebook, we will learn to use groupby by analyzing sea-surface temperature data.
import numpy as np
import pandas as pd
import xarray as xr
from matplotlib import pyplot as plt
#%config InlineBackend.figure_format = 'retina'
plt.ion() # To trigger the interactive inline mode
plt.rcParams['figure.figsize'] = (6,5)
First we load a dataset. We will use the NOAA Extended Reconstructed Sea Surface Temperature (ERSST) v5 product, a widely used and trusted gridded compilation of of historical data going back to 1854.
Since the data is provided via an OPeNDAP server, we can load it directly without downloading anything:
url = 'http://www.esrl.noaa.gov/psd/thredds/dodsC/Datasets/noaa.ersst.v5/sst.mnmean.nc'
ds = xr.open_dataset(url, drop_variables=['time_bnds'])
ds = ds.sel(time=slice('1960', '2018')).load()
ds
<xarray.Dataset>
Dimensions: (lat: 89, lon: 180, time: 708)
Coordinates:
* lat (lat) float32 88.0 86.0 84.0 82.0 80.0 ... -82.0 -84.0 -86.0 -88.0
* lon (lon) float32 0.0 2.0 4.0 6.0 8.0 ... 350.0 352.0 354.0 356.0 358.0
* time (time) datetime64[ns] 1960-01-01 1960-02-01 ... 2018-12-01
Data variables:
sst (time, lat, lon) float32 -1.8 -1.8 -1.8 -1.8 ... nan nan nan nan
Attributes: (12/38)
climatology: Climatology is based on 1971-2000 SST, X...
description: In situ data: ICOADS2.5 before 2007 and ...
keywords_vocabulary: NASA Global Change Master Directory (GCM...
keywords: Earth Science > Oceans > Ocean Temperatu...
instrument: Conventional thermometers
source_comment: SSTs were observed by conventional therm...
... ...
license: No constraints on data access or use
comment: SSTs were observed by conventional therm...
summary: ERSST.v5 is developed based on v4 after ...
dataset_title: NOAA Extended Reconstructed SST V5
data_modified: 2021-03-07
DODS_EXTRA.Unlimited_Dimension: time- lat: 89
- lon: 180
- time: 708
- lat(lat)float3288.0 86.0 84.0 ... -86.0 -88.0
- units :
- degrees_north
- long_name :
- Latitude
- actual_range :
- [ 88. -88.]
- standard_name :
- latitude
- axis :
- Y
- coordinate_defines :
- center
array([ 88., 86., 84., 82., 80., 78., 76., 74., 72., 70., 68., 66., 64., 62., 60., 58., 56., 54., 52., 50., 48., 46., 44., 42., 40., 38., 36., 34., 32., 30., 28., 26., 24., 22., 20., 18., 16., 14., 12., 10., 8., 6., 4., 2., 0., -2., -4., -6., -8., -10., -12., -14., -16., -18., -20., -22., -24., -26., -28., -30., -32., -34., -36., -38., -40., -42., -44., -46., -48., -50., -52., -54., -56., -58., -60., -62., -64., -66., -68., -70., -72., -74., -76., -78., -80., -82., -84., -86., -88.], dtype=float32) - lon(lon)float320.0 2.0 4.0 ... 354.0 356.0 358.0
- units :
- degrees_east
- long_name :
- Longitude
- actual_range :
- [ 0. 358.]
- standard_name :
- longitude
- axis :
- X
- coordinate_defines :
- center
array([ 0., 2., 4., 6., 8., 10., 12., 14., 16., 18., 20., 22., 24., 26., 28., 30., 32., 34., 36., 38., 40., 42., 44., 46., 48., 50., 52., 54., 56., 58., 60., 62., 64., 66., 68., 70., 72., 74., 76., 78., 80., 82., 84., 86., 88., 90., 92., 94., 96., 98., 100., 102., 104., 106., 108., 110., 112., 114., 116., 118., 120., 122., 124., 126., 128., 130., 132., 134., 136., 138., 140., 142., 144., 146., 148., 150., 152., 154., 156., 158., 160., 162., 164., 166., 168., 170., 172., 174., 176., 178., 180., 182., 184., 186., 188., 190., 192., 194., 196., 198., 200., 202., 204., 206., 208., 210., 212., 214., 216., 218., 220., 222., 224., 226., 228., 230., 232., 234., 236., 238., 240., 242., 244., 246., 248., 250., 252., 254., 256., 258., 260., 262., 264., 266., 268., 270., 272., 274., 276., 278., 280., 282., 284., 286., 288., 290., 292., 294., 296., 298., 300., 302., 304., 306., 308., 310., 312., 314., 316., 318., 320., 322., 324., 326., 328., 330., 332., 334., 336., 338., 340., 342., 344., 346., 348., 350., 352., 354., 356., 358.], dtype=float32) - time(time)datetime64[ns]1960-01-01 ... 2018-12-01
- long_name :
- Time
- delta_t :
- 0000-01-00 00:00:00
- avg_period :
- 0000-01-00 00:00:00
- prev_avg_period :
- 0000-00-07 00:00:00
- standard_name :
- time
- axis :
- T
- actual_range :
- [19723. 80750.]
- _ChunkSizes :
- 1
array(['1960-01-01T00:00:00.000000000', '1960-02-01T00:00:00.000000000', '1960-03-01T00:00:00.000000000', ..., '2018-10-01T00:00:00.000000000', '2018-11-01T00:00:00.000000000', '2018-12-01T00:00:00.000000000'], dtype='datetime64[ns]')
- sst(time, lat, lon)float32-1.8 -1.8 -1.8 -1.8 ... nan nan nan
- long_name :
- Monthly Means of Sea Surface Temperature
- units :
- degC
- var_desc :
- Sea Surface Temperature
- level_desc :
- Surface
- statistic :
- Mean
- dataset :
- NOAA Extended Reconstructed SST V5
- parent_stat :
- Individual Values
- actual_range :
- [-1.8 42.32636]
- valid_range :
- [-1.8 45. ]
- _ChunkSizes :
- [ 1 89 180]
array([[[-1.8, -1.8, -1.8, ..., -1.8, -1.8, -1.8], [-1.8, -1.8, -1.8, ..., -1.8, -1.8, -1.8], [-1.8, -1.8, -1.8, ..., -1.8, -1.8, -1.8], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[-1.8, -1.8, -1.8, ..., -1.8, -1.8, -1.8], [-1.8, -1.8, -1.8, ..., -1.8, -1.8, -1.8], [-1.8, -1.8, -1.8, ..., -1.8, -1.8, -1.8], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[-1.8, -1.8, -1.8, ..., -1.8, -1.8, -1.8], [-1.8, -1.8, -1.8, ..., -1.8, -1.8, -1.8], [-1.8, -1.8, -1.8, ..., -1.8, -1.8, -1.8], ..., ... ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[-1.8, -1.8, -1.8, ..., -1.8, -1.8, -1.8], [-1.8, -1.8, -1.8, ..., -1.8, -1.8, -1.8], [-1.8, -1.8, -1.8, ..., -1.8, -1.8, -1.8], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[-1.8, -1.8, -1.8, ..., -1.8, -1.8, -1.8], [-1.8, -1.8, -1.8, ..., -1.8, -1.8, -1.8], [-1.8, -1.8, -1.8, ..., -1.8, -1.8, -1.8], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]], dtype=float32)
- climatology :
- Climatology is based on 1971-2000 SST, Xue, Y., T. M. Smith, and R. W. Reynolds, 2003: Interdecadal changes of 30-yr SST normals during 1871.2000. Journal of Climate, 16, 1601-1612.
- description :
- In situ data: ICOADS2.5 before 2007 and NCEP in situ data from 2008 to present. Ice data: HadISST ice before 2010 and NCEP ice after 2010.
- keywords_vocabulary :
- NASA Global Change Master Directory (GCMD) Science Keywords
- keywords :
- Earth Science > Oceans > Ocean Temperature > Sea Surface Temperature >
- instrument :
- Conventional thermometers
- source_comment :
- SSTs were observed by conventional thermometers in Buckets (insulated or un-insulated canvas and wooded buckets) or Engine Room Intaker
- geospatial_lon_min :
- -1.0
- geospatial_lon_max :
- 359.0
- geospatial_laty_max :
- 89.0
- geospatial_laty_min :
- -89.0
- geospatial_lat_max :
- 89.0
- geospatial_lat_min :
- -89.0
- geospatial_lat_units :
- degrees_north
- geospatial_lon_units :
- degrees_east
- cdm_data_type :
- Grid
- project :
- NOAA Extended Reconstructed Sea Surface Temperature (ERSST)
- original_publisher_url :
- http://www.ncdc.noaa.gov
- References :
- https://www.ncdc.noaa.gov/data-access/marineocean-data/extended-reconstructed-sea-surface-temperature-ersst-v5 at NCEI and http://www.esrl.noaa.gov/psd/data/gridded/data.noaa.ersst.v5.html
- source :
- In situ data: ICOADS R3.0 before 2015, NCEP in situ GTS from 2016 to present, and Argo SST from 1999 to present. Ice data: HadISST2 ice before 2015, and NCEP ice after 2015
- title :
- NOAA ERSSTv5 (in situ only)
- history :
- created 07/2017 by PSD data using NCEI's ERSST V5 NetCDF values
- institution :
- This version written at NOAA/ESRL PSD: obtained from NOAA/NESDIS/National Centers for Environmental Information and time aggregated. Original Full Source: NOAA/NESDIS/NCEI/CCOG
- citation :
- Huang et al, 2017: Extended Reconstructed Sea Surface Temperatures Version 5 (ERSSTv5): Upgrades, Validations, and Intercomparisons. Journal of Climate, https://doi.org/10.1175/JCLI-D-16-0836.1
- platform :
- Ship and Buoy SSTs from ICOADS R3.0 and NCEP GTS
- standard_name_vocabulary :
- CF Standard Name Table (v40, 25 January 2017)
- processing_level :
- NOAA Level 4
- Conventions :
- CF-1.6, ACDD-1.3
- metadata_link :
- :metadata_link = https://doi.org/10.7289/V5T72FNM (original format)
- creator_name :
- Boyin Huang (original)
- date_created :
- 2017-06-30T12:18:00Z (original)
- product_version :
- Version 5
- creator_url_original :
- https://www.ncei.noaa.gov
- license :
- No constraints on data access or use
- comment :
- SSTs were observed by conventional thermometers in Buckets (insulated or un-insulated canvas and wooded buckets), Engine Room Intakers, or floats and drifters
- summary :
- ERSST.v5 is developed based on v4 after revisions of 8 parameters using updated data sets and advanced knowledge of ERSST analysis
- dataset_title :
- NOAA Extended Reconstructed SST V5
- data_modified :
- 2021-03-07
- DODS_EXTRA.Unlimited_Dimension :
- time
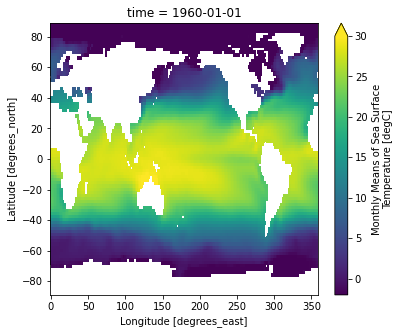
Let’s do some basic visualizations of the data, just to make sure it looks reasonable.
ds.sst[0].plot(vmin=-2, vmax=30)
<matplotlib.collections.QuadMesh at 0x7f007cbfe8b0>
Note that xarray correctly parsed the time index, resulting in a Pandas datetime index on the time dimension.
ds.time
<xarray.DataArray 'time' (time: 708)>
array(['1960-01-01T00:00:00.000000000', '1960-02-01T00:00:00.000000000',
'1960-03-01T00:00:00.000000000', ..., '2018-10-01T00:00:00.000000000',
'2018-11-01T00:00:00.000000000', '2018-12-01T00:00:00.000000000'],
dtype='datetime64[ns]')
Coordinates:
* time (time) datetime64[ns] 1960-01-01 1960-02-01 ... 2018-12-01
Attributes:
long_name: Time
delta_t: 0000-01-00 00:00:00
avg_period: 0000-01-00 00:00:00
prev_avg_period: 0000-00-07 00:00:00
standard_name: time
axis: T
actual_range: [19723. 80750.]
_ChunkSizes: 1- time: 708
- 1960-01-01 1960-02-01 1960-03-01 ... 2018-10-01 2018-11-01 2018-12-01
array(['1960-01-01T00:00:00.000000000', '1960-02-01T00:00:00.000000000', '1960-03-01T00:00:00.000000000', ..., '2018-10-01T00:00:00.000000000', '2018-11-01T00:00:00.000000000', '2018-12-01T00:00:00.000000000'], dtype='datetime64[ns]') - time(time)datetime64[ns]1960-01-01 ... 2018-12-01
- long_name :
- Time
- delta_t :
- 0000-01-00 00:00:00
- avg_period :
- 0000-01-00 00:00:00
- prev_avg_period :
- 0000-00-07 00:00:00
- standard_name :
- time
- axis :
- T
- actual_range :
- [19723. 80750.]
- _ChunkSizes :
- 1
array(['1960-01-01T00:00:00.000000000', '1960-02-01T00:00:00.000000000', '1960-03-01T00:00:00.000000000', ..., '2018-10-01T00:00:00.000000000', '2018-11-01T00:00:00.000000000', '2018-12-01T00:00:00.000000000'], dtype='datetime64[ns]')
- long_name :
- Time
- delta_t :
- 0000-01-00 00:00:00
- avg_period :
- 0000-01-00 00:00:00
- prev_avg_period :
- 0000-00-07 00:00:00
- standard_name :
- time
- axis :
- T
- actual_range :
- [19723. 80750.]
- _ChunkSizes :
- 1
ds.sst.sel(lon=300, lat=50).plot()
[<matplotlib.lines.Line2D at 0x7f007cb2b730>]
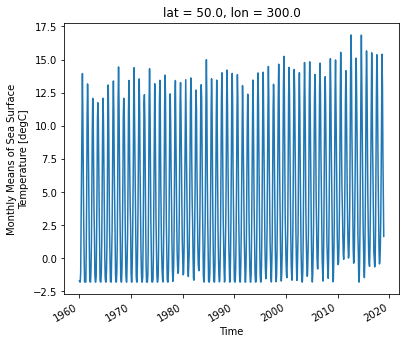
As we can see from the plot, the timeseries at any one point is totally dominated by the seasonal cycle. We would like to remove this seasonal cycle (called the “climatology”) in order to better see the long-term variaitions in temperature. We will accomplish this using groupby.
The syntax of Xarray’s groupby is almost identical to Pandas.
help(ds.groupby)
Help on method groupby in module xarray.core.common:
groupby(group, squeeze: bool = True, restore_coord_dims: bool = None) method of xarray.core.dataset.Dataset instance
Returns a GroupBy object for performing grouped operations.
Parameters
----------
group : str, DataArray or IndexVariable
Array whose unique values should be used to group this array. If a
string, must be the name of a variable contained in this dataset.
squeeze : bool, optional
If "group" is a dimension of any arrays in this dataset, `squeeze`
controls whether the subarrays have a dimension of length 1 along
that dimension or if the dimension is squeezed out.
restore_coord_dims : bool, optional
If True, also restore the dimension order of multi-dimensional
coordinates.
Returns
-------
grouped
A `GroupBy` object patterned after `pandas.GroupBy` that can be
iterated over in the form of `(unique_value, grouped_array)` pairs.
Examples
--------
Calculate daily anomalies for daily data:
>>> da = xr.DataArray(
... np.linspace(0, 1826, num=1827),
... coords=[pd.date_range("1/1/2000", "31/12/2004", freq="D")],
... dims="time",
... )
>>> da
<xarray.DataArray (time: 1827)>
array([0.000e+00, 1.000e+00, 2.000e+00, ..., 1.824e+03, 1.825e+03,
1.826e+03])
Coordinates:
* time (time) datetime64[ns] 2000-01-01 2000-01-02 ... 2004-12-31
>>> da.groupby("time.dayofyear") - da.groupby("time.dayofyear").mean("time")
<xarray.DataArray (time: 1827)>
array([-730.8, -730.8, -730.8, ..., 730.2, 730.2, 730.5])
Coordinates:
* time (time) datetime64[ns] 2000-01-01 2000-01-02 ... 2004-12-31
dayofyear (time) int64 1 2 3 4 5 6 7 8 ... 359 360 361 362 363 364 365 366
See Also
--------
core.groupby.DataArrayGroupBy
core.groupby.DatasetGroupBy
Split Step¶
The most important argument is group: this defines the unique values we will use to “split” the data for grouped analysis. We can pass either a DataArray or a name of a variable in the dataset. Lets first use a DataArray. Just like with Pandas, we can use the time index to extract specific components of dates and times. Xarray uses a special syntax for this .dt, called the DatetimeAccessor.
ds.time.dt
<xarray.core.accessor_dt.DatetimeAccessor at 0x7f007caa0610>
ds.time.dt.month
<xarray.DataArray 'month' (time: 708)>
array([ 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5,
6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10,
11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3,
4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8,
9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1,
2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6,
7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11,
12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4,
5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9,
10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2,
3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7,
8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12,
1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5,
6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10,
11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3,
4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8,
9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1,
2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6,
7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11,
12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4,
...
3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7,
8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12,
1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5,
6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10,
11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3,
4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8,
9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1,
2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6,
7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11,
12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4,
5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9,
10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2,
3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7,
8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12,
1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5,
6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10,
11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3,
4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8,
9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1,
2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12])
Coordinates:
* time (time) datetime64[ns] 1960-01-01 1960-02-01 ... 2018-12-01- time: 708
- 1 2 3 4 5 6 7 8 9 10 11 12 1 2 3 ... 11 12 1 2 3 4 5 6 7 8 9 10 11 12
array([ 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, ... 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12]) - time(time)datetime64[ns]1960-01-01 ... 2018-12-01
- long_name :
- Time
- delta_t :
- 0000-01-00 00:00:00
- avg_period :
- 0000-01-00 00:00:00
- prev_avg_period :
- 0000-00-07 00:00:00
- standard_name :
- time
- axis :
- T
- actual_range :
- [19723. 80750.]
- _ChunkSizes :
- 1
array(['1960-01-01T00:00:00.000000000', '1960-02-01T00:00:00.000000000', '1960-03-01T00:00:00.000000000', ..., '2018-10-01T00:00:00.000000000', '2018-11-01T00:00:00.000000000', '2018-12-01T00:00:00.000000000'], dtype='datetime64[ns]')
ds.time.dt.year
We can use these arrays in a groupby operation:
gb = ds.groupby(ds.time.dt.month)
gb
DatasetGroupBy, grouped over 'month'
12 groups with labels 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12.
Xarray also offers a more concise syntax when the variable you’re grouping on is already present in the dataset. This is identical to the previous line:
gb = ds.groupby('time.month')
gb
DatasetGroupBy, grouped over 'month'
12 groups with labels 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12.
Now that the data are split, we can manually iterate over the group. The iterator returns the key (group name) and the value (the actual dataset corresponding to that group) for each group.
for group_name, group_ds in gb:
# stop iterating after the first loop
break
print(group_name)
group_ds
1
<xarray.Dataset>
Dimensions: (lat: 89, lon: 180, time: 59)
Coordinates:
* lat (lat) float32 88.0 86.0 84.0 82.0 80.0 ... -82.0 -84.0 -86.0 -88.0
* lon (lon) float32 0.0 2.0 4.0 6.0 8.0 ... 350.0 352.0 354.0 356.0 358.0
* time (time) datetime64[ns] 1960-01-01 1961-01-01 ... 2018-01-01
Data variables:
sst (time, lat, lon) float32 -1.8 -1.8 -1.8 -1.8 ... nan nan nan nan
Attributes: (12/38)
climatology: Climatology is based on 1971-2000 SST, X...
description: In situ data: ICOADS2.5 before 2007 and ...
keywords_vocabulary: NASA Global Change Master Directory (GCM...
keywords: Earth Science > Oceans > Ocean Temperatu...
instrument: Conventional thermometers
source_comment: SSTs were observed by conventional therm...
... ...
license: No constraints on data access or use
comment: SSTs were observed by conventional therm...
summary: ERSST.v5 is developed based on v4 after ...
dataset_title: NOAA Extended Reconstructed SST V5
data_modified: 2021-03-07
DODS_EXTRA.Unlimited_Dimension: time- lat: 89
- lon: 180
- time: 59
- lat(lat)float3288.0 86.0 84.0 ... -86.0 -88.0
- units :
- degrees_north
- long_name :
- Latitude
- actual_range :
- [ 88. -88.]
- standard_name :
- latitude
- axis :
- Y
- coordinate_defines :
- center
array([ 88., 86., 84., 82., 80., 78., 76., 74., 72., 70., 68., 66., 64., 62., 60., 58., 56., 54., 52., 50., 48., 46., 44., 42., 40., 38., 36., 34., 32., 30., 28., 26., 24., 22., 20., 18., 16., 14., 12., 10., 8., 6., 4., 2., 0., -2., -4., -6., -8., -10., -12., -14., -16., -18., -20., -22., -24., -26., -28., -30., -32., -34., -36., -38., -40., -42., -44., -46., -48., -50., -52., -54., -56., -58., -60., -62., -64., -66., -68., -70., -72., -74., -76., -78., -80., -82., -84., -86., -88.], dtype=float32) - lon(lon)float320.0 2.0 4.0 ... 354.0 356.0 358.0
- units :
- degrees_east
- long_name :
- Longitude
- actual_range :
- [ 0. 358.]
- standard_name :
- longitude
- axis :
- X
- coordinate_defines :
- center
array([ 0., 2., 4., 6., 8., 10., 12., 14., 16., 18., 20., 22., 24., 26., 28., 30., 32., 34., 36., 38., 40., 42., 44., 46., 48., 50., 52., 54., 56., 58., 60., 62., 64., 66., 68., 70., 72., 74., 76., 78., 80., 82., 84., 86., 88., 90., 92., 94., 96., 98., 100., 102., 104., 106., 108., 110., 112., 114., 116., 118., 120., 122., 124., 126., 128., 130., 132., 134., 136., 138., 140., 142., 144., 146., 148., 150., 152., 154., 156., 158., 160., 162., 164., 166., 168., 170., 172., 174., 176., 178., 180., 182., 184., 186., 188., 190., 192., 194., 196., 198., 200., 202., 204., 206., 208., 210., 212., 214., 216., 218., 220., 222., 224., 226., 228., 230., 232., 234., 236., 238., 240., 242., 244., 246., 248., 250., 252., 254., 256., 258., 260., 262., 264., 266., 268., 270., 272., 274., 276., 278., 280., 282., 284., 286., 288., 290., 292., 294., 296., 298., 300., 302., 304., 306., 308., 310., 312., 314., 316., 318., 320., 322., 324., 326., 328., 330., 332., 334., 336., 338., 340., 342., 344., 346., 348., 350., 352., 354., 356., 358.], dtype=float32) - time(time)datetime64[ns]1960-01-01 ... 2018-01-01
- long_name :
- Time
- delta_t :
- 0000-01-00 00:00:00
- avg_period :
- 0000-01-00 00:00:00
- prev_avg_period :
- 0000-00-07 00:00:00
- standard_name :
- time
- axis :
- T
- actual_range :
- [19723. 80750.]
- _ChunkSizes :
- 1
array(['1960-01-01T00:00:00.000000000', '1961-01-01T00:00:00.000000000', '1962-01-01T00:00:00.000000000', '1963-01-01T00:00:00.000000000', '1964-01-01T00:00:00.000000000', '1965-01-01T00:00:00.000000000', '1966-01-01T00:00:00.000000000', '1967-01-01T00:00:00.000000000', '1968-01-01T00:00:00.000000000', '1969-01-01T00:00:00.000000000', '1970-01-01T00:00:00.000000000', '1971-01-01T00:00:00.000000000', '1972-01-01T00:00:00.000000000', '1973-01-01T00:00:00.000000000', '1974-01-01T00:00:00.000000000', '1975-01-01T00:00:00.000000000', '1976-01-01T00:00:00.000000000', '1977-01-01T00:00:00.000000000', '1978-01-01T00:00:00.000000000', '1979-01-01T00:00:00.000000000', '1980-01-01T00:00:00.000000000', '1981-01-01T00:00:00.000000000', '1982-01-01T00:00:00.000000000', '1983-01-01T00:00:00.000000000', '1984-01-01T00:00:00.000000000', '1985-01-01T00:00:00.000000000', '1986-01-01T00:00:00.000000000', '1987-01-01T00:00:00.000000000', '1988-01-01T00:00:00.000000000', '1989-01-01T00:00:00.000000000', '1990-01-01T00:00:00.000000000', '1991-01-01T00:00:00.000000000', '1992-01-01T00:00:00.000000000', '1993-01-01T00:00:00.000000000', '1994-01-01T00:00:00.000000000', '1995-01-01T00:00:00.000000000', '1996-01-01T00:00:00.000000000', '1997-01-01T00:00:00.000000000', '1998-01-01T00:00:00.000000000', '1999-01-01T00:00:00.000000000', '2000-01-01T00:00:00.000000000', '2001-01-01T00:00:00.000000000', '2002-01-01T00:00:00.000000000', '2003-01-01T00:00:00.000000000', '2004-01-01T00:00:00.000000000', '2005-01-01T00:00:00.000000000', '2006-01-01T00:00:00.000000000', '2007-01-01T00:00:00.000000000', '2008-01-01T00:00:00.000000000', '2009-01-01T00:00:00.000000000', '2010-01-01T00:00:00.000000000', '2011-01-01T00:00:00.000000000', '2012-01-01T00:00:00.000000000', '2013-01-01T00:00:00.000000000', '2014-01-01T00:00:00.000000000', '2015-01-01T00:00:00.000000000', '2016-01-01T00:00:00.000000000', '2017-01-01T00:00:00.000000000', '2018-01-01T00:00:00.000000000'], dtype='datetime64[ns]')
- sst(time, lat, lon)float32-1.8 -1.8 -1.8 -1.8 ... nan nan nan
- long_name :
- Monthly Means of Sea Surface Temperature
- units :
- degC
- var_desc :
- Sea Surface Temperature
- level_desc :
- Surface
- statistic :
- Mean
- dataset :
- NOAA Extended Reconstructed SST V5
- parent_stat :
- Individual Values
- actual_range :
- [-1.8 42.32636]
- valid_range :
- [-1.8 45. ]
- _ChunkSizes :
- [ 1 89 180]
array([[[-1.8, -1.8, -1.8, ..., -1.8, -1.8, -1.8], [-1.8, -1.8, -1.8, ..., -1.8, -1.8, -1.8], [-1.8, -1.8, -1.8, ..., -1.8, -1.8, -1.8], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[-1.8, -1.8, -1.8, ..., -1.8, -1.8, -1.8], [-1.8, -1.8, -1.8, ..., -1.8, -1.8, -1.8], [-1.8, -1.8, -1.8, ..., -1.8, -1.8, -1.8], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[-1.8, -1.8, -1.8, ..., -1.8, -1.8, -1.8], [-1.8, -1.8, -1.8, ..., -1.8, -1.8, -1.8], [-1.8, -1.8, -1.8, ..., -1.8, -1.8, -1.8], ..., ... ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[-1.8, -1.8, -1.8, ..., -1.8, -1.8, -1.8], [-1.8, -1.8, -1.8, ..., -1.8, -1.8, -1.8], [-1.8, -1.8, -1.8, ..., -1.8, -1.8, -1.8], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[-1.8, -1.8, -1.8, ..., -1.8, -1.8, -1.8], [-1.8, -1.8, -1.8, ..., -1.8, -1.8, -1.8], [-1.8, -1.8, -1.8, ..., -1.8, -1.8, -1.8], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]], dtype=float32)
- climatology :
- Climatology is based on 1971-2000 SST, Xue, Y., T. M. Smith, and R. W. Reynolds, 2003: Interdecadal changes of 30-yr SST normals during 1871.2000. Journal of Climate, 16, 1601-1612.
- description :
- In situ data: ICOADS2.5 before 2007 and NCEP in situ data from 2008 to present. Ice data: HadISST ice before 2010 and NCEP ice after 2010.
- keywords_vocabulary :
- NASA Global Change Master Directory (GCMD) Science Keywords
- keywords :
- Earth Science > Oceans > Ocean Temperature > Sea Surface Temperature >
- instrument :
- Conventional thermometers
- source_comment :
- SSTs were observed by conventional thermometers in Buckets (insulated or un-insulated canvas and wooded buckets) or Engine Room Intaker
- geospatial_lon_min :
- -1.0
- geospatial_lon_max :
- 359.0
- geospatial_laty_max :
- 89.0
- geospatial_laty_min :
- -89.0
- geospatial_lat_max :
- 89.0
- geospatial_lat_min :
- -89.0
- geospatial_lat_units :
- degrees_north
- geospatial_lon_units :
- degrees_east
- cdm_data_type :
- Grid
- project :
- NOAA Extended Reconstructed Sea Surface Temperature (ERSST)
- original_publisher_url :
- http://www.ncdc.noaa.gov
- References :
- https://www.ncdc.noaa.gov/data-access/marineocean-data/extended-reconstructed-sea-surface-temperature-ersst-v5 at NCEI and http://www.esrl.noaa.gov/psd/data/gridded/data.noaa.ersst.v5.html
- source :
- In situ data: ICOADS R3.0 before 2015, NCEP in situ GTS from 2016 to present, and Argo SST from 1999 to present. Ice data: HadISST2 ice before 2015, and NCEP ice after 2015
- title :
- NOAA ERSSTv5 (in situ only)
- history :
- created 07/2017 by PSD data using NCEI's ERSST V5 NetCDF values
- institution :
- This version written at NOAA/ESRL PSD: obtained from NOAA/NESDIS/National Centers for Environmental Information and time aggregated. Original Full Source: NOAA/NESDIS/NCEI/CCOG
- citation :
- Huang et al, 2017: Extended Reconstructed Sea Surface Temperatures Version 5 (ERSSTv5): Upgrades, Validations, and Intercomparisons. Journal of Climate, https://doi.org/10.1175/JCLI-D-16-0836.1
- platform :
- Ship and Buoy SSTs from ICOADS R3.0 and NCEP GTS
- standard_name_vocabulary :
- CF Standard Name Table (v40, 25 January 2017)
- processing_level :
- NOAA Level 4
- Conventions :
- CF-1.6, ACDD-1.3
- metadata_link :
- :metadata_link = https://doi.org/10.7289/V5T72FNM (original format)
- creator_name :
- Boyin Huang (original)
- date_created :
- 2017-06-30T12:18:00Z (original)
- product_version :
- Version 5
- creator_url_original :
- https://www.ncei.noaa.gov
- license :
- No constraints on data access or use
- comment :
- SSTs were observed by conventional thermometers in Buckets (insulated or un-insulated canvas and wooded buckets), Engine Room Intakers, or floats and drifters
- summary :
- ERSST.v5 is developed based on v4 after revisions of 8 parameters using updated data sets and advanced knowledge of ERSST analysis
- dataset_title :
- NOAA Extended Reconstructed SST V5
- data_modified :
- 2021-03-07
- DODS_EXTRA.Unlimited_Dimension :
- time
Apply & Combine¶
Now that we have groups defined, it’s time to “apply” a calculation to the group. Like in Pandas, these calculations can either be:
aggregation: reduces the size of the group
transformation: preserves the group’s full size
At then end of the apply step, xarray will automatically combine the aggregated / transformed groups back into a single object.
The most fundamental way to apply is with the .apply method.
help(gb.apply)
Help on method apply in module xarray.core.groupby:
apply(func, args=(), shortcut=None, **kwargs) method of xarray.core.groupby.DatasetGroupBy instance
Backward compatible implementation of ``map``
See Also
--------
DatasetGroupBy.map
Aggregations¶
.apply accepts as its argument a function. We can pass an existing function:
gb.apply(np.mean)
<xarray.Dataset>
Dimensions: (month: 12)
Coordinates:
* month (month) int64 1 2 3 4 5 6 7 8 9 10 11 12
Data variables:
sst (month) float32 13.66 13.77 13.76 13.68 ... 13.98 13.69 13.51 13.53- month: 12
- month(month)int641 2 3 4 5 6 7 8 9 10 11 12
array([ 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12])
- sst(month)float3213.66 13.77 13.76 ... 13.51 13.53
array([13.659641, 13.768647, 13.76488 , 13.684034, 13.642146, 13.713043, 13.921847, 14.093956, 13.982147, 13.691116, 13.506494, 13.529454], dtype=float32)
Because we specified no extra arguments (like axis) the function was applied over all space and time dimensions. This is not what we wanted. Instead, we could define a custom function. This function takes a single argument–the group dataset–and returns a new dataset to be combined:
def time_mean(a):
return a.mean(dim='time')
gb.apply(time_mean)
<xarray.Dataset>
Dimensions: (lat: 89, lon: 180, month: 12)
Coordinates:
* lat (lat) float32 88.0 86.0 84.0 82.0 80.0 ... -82.0 -84.0 -86.0 -88.0
* lon (lon) float32 0.0 2.0 4.0 6.0 8.0 ... 350.0 352.0 354.0 356.0 358.0
* month (month) int64 1 2 3 4 5 6 7 8 9 10 11 12
Data variables:
sst (month, lat, lon) float32 -1.8 -1.8 -1.8 -1.8 ... nan nan nan nan- lat: 89
- lon: 180
- month: 12
- lat(lat)float3288.0 86.0 84.0 ... -86.0 -88.0
- units :
- degrees_north
- long_name :
- Latitude
- actual_range :
- [ 88. -88.]
- standard_name :
- latitude
- axis :
- Y
- coordinate_defines :
- center
array([ 88., 86., 84., 82., 80., 78., 76., 74., 72., 70., 68., 66., 64., 62., 60., 58., 56., 54., 52., 50., 48., 46., 44., 42., 40., 38., 36., 34., 32., 30., 28., 26., 24., 22., 20., 18., 16., 14., 12., 10., 8., 6., 4., 2., 0., -2., -4., -6., -8., -10., -12., -14., -16., -18., -20., -22., -24., -26., -28., -30., -32., -34., -36., -38., -40., -42., -44., -46., -48., -50., -52., -54., -56., -58., -60., -62., -64., -66., -68., -70., -72., -74., -76., -78., -80., -82., -84., -86., -88.], dtype=float32) - lon(lon)float320.0 2.0 4.0 ... 354.0 356.0 358.0
- units :
- degrees_east
- long_name :
- Longitude
- actual_range :
- [ 0. 358.]
- standard_name :
- longitude
- axis :
- X
- coordinate_defines :
- center
array([ 0., 2., 4., 6., 8., 10., 12., 14., 16., 18., 20., 22., 24., 26., 28., 30., 32., 34., 36., 38., 40., 42., 44., 46., 48., 50., 52., 54., 56., 58., 60., 62., 64., 66., 68., 70., 72., 74., 76., 78., 80., 82., 84., 86., 88., 90., 92., 94., 96., 98., 100., 102., 104., 106., 108., 110., 112., 114., 116., 118., 120., 122., 124., 126., 128., 130., 132., 134., 136., 138., 140., 142., 144., 146., 148., 150., 152., 154., 156., 158., 160., 162., 164., 166., 168., 170., 172., 174., 176., 178., 180., 182., 184., 186., 188., 190., 192., 194., 196., 198., 200., 202., 204., 206., 208., 210., 212., 214., 216., 218., 220., 222., 224., 226., 228., 230., 232., 234., 236., 238., 240., 242., 244., 246., 248., 250., 252., 254., 256., 258., 260., 262., 264., 266., 268., 270., 272., 274., 276., 278., 280., 282., 284., 286., 288., 290., 292., 294., 296., 298., 300., 302., 304., 306., 308., 310., 312., 314., 316., 318., 320., 322., 324., 326., 328., 330., 332., 334., 336., 338., 340., 342., 344., 346., 348., 350., 352., 354., 356., 358.], dtype=float32) - month(month)int641 2 3 4 5 6 7 8 9 10 11 12
array([ 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12])
- sst(month, lat, lon)float32-1.8 -1.8 -1.8 -1.8 ... nan nan nan
array([[[-1.8000009, -1.8000009, -1.8000009, ..., -1.8000009, -1.8000009, -1.8000009], [-1.8000009, -1.8000009, -1.8000009, ..., -1.8000009, -1.8000009, -1.8000009], [-1.8000009, -1.8000009, -1.8000009, ..., -1.8000009, -1.8000009, -1.8000009], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[-1.8000009, -1.8000009, -1.8000009, ..., -1.8000009, -1.8000009, -1.8000009], [-1.8000009, -1.8000009, -1.8000009, ..., -1.8000009, -1.8000009, -1.8000009], [-1.8000009, -1.8000009, -1.8000009, ..., -1.8000009, -1.8000009, -1.8000009], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[-1.7995025, -1.7995973, -1.7998415, ..., -1.7997988, -1.7996519, -1.7995045], [-1.7995876, -1.7997634, -1.8000009, ..., -1.8000009, -1.7998358, -1.7996247], [-1.8000009, -1.8000009, -1.8000009, ..., -1.8000009, -1.8000009, -1.8000009], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]], dtype=float32)
Like Pandas, xarray’s groupby object has many built-in aggregation operations (e.g. mean, min, max, std, etc):
# this does the same thing as the previous cell
ds_mm = gb.mean(dim='time')
ds_mm
<xarray.Dataset>
Dimensions: (lat: 89, lon: 180, month: 12)
Coordinates:
* lat (lat) float32 88.0 86.0 84.0 82.0 80.0 ... -82.0 -84.0 -86.0 -88.0
* lon (lon) float32 0.0 2.0 4.0 6.0 8.0 ... 350.0 352.0 354.0 356.0 358.0
* month (month) int64 1 2 3 4 5 6 7 8 9 10 11 12
Data variables:
sst (month, lat, lon) float32 -1.8 -1.8 -1.8 -1.8 ... nan nan nan nan- lat: 89
- lon: 180
- month: 12
- lat(lat)float3288.0 86.0 84.0 ... -86.0 -88.0
- units :
- degrees_north
- long_name :
- Latitude
- actual_range :
- [ 88. -88.]
- standard_name :
- latitude
- axis :
- Y
- coordinate_defines :
- center
array([ 88., 86., 84., 82., 80., 78., 76., 74., 72., 70., 68., 66., 64., 62., 60., 58., 56., 54., 52., 50., 48., 46., 44., 42., 40., 38., 36., 34., 32., 30., 28., 26., 24., 22., 20., 18., 16., 14., 12., 10., 8., 6., 4., 2., 0., -2., -4., -6., -8., -10., -12., -14., -16., -18., -20., -22., -24., -26., -28., -30., -32., -34., -36., -38., -40., -42., -44., -46., -48., -50., -52., -54., -56., -58., -60., -62., -64., -66., -68., -70., -72., -74., -76., -78., -80., -82., -84., -86., -88.], dtype=float32) - lon(lon)float320.0 2.0 4.0 ... 354.0 356.0 358.0
- units :
- degrees_east
- long_name :
- Longitude
- actual_range :
- [ 0. 358.]
- standard_name :
- longitude
- axis :
- X
- coordinate_defines :
- center
array([ 0., 2., 4., 6., 8., 10., 12., 14., 16., 18., 20., 22., 24., 26., 28., 30., 32., 34., 36., 38., 40., 42., 44., 46., 48., 50., 52., 54., 56., 58., 60., 62., 64., 66., 68., 70., 72., 74., 76., 78., 80., 82., 84., 86., 88., 90., 92., 94., 96., 98., 100., 102., 104., 106., 108., 110., 112., 114., 116., 118., 120., 122., 124., 126., 128., 130., 132., 134., 136., 138., 140., 142., 144., 146., 148., 150., 152., 154., 156., 158., 160., 162., 164., 166., 168., 170., 172., 174., 176., 178., 180., 182., 184., 186., 188., 190., 192., 194., 196., 198., 200., 202., 204., 206., 208., 210., 212., 214., 216., 218., 220., 222., 224., 226., 228., 230., 232., 234., 236., 238., 240., 242., 244., 246., 248., 250., 252., 254., 256., 258., 260., 262., 264., 266., 268., 270., 272., 274., 276., 278., 280., 282., 284., 286., 288., 290., 292., 294., 296., 298., 300., 302., 304., 306., 308., 310., 312., 314., 316., 318., 320., 322., 324., 326., 328., 330., 332., 334., 336., 338., 340., 342., 344., 346., 348., 350., 352., 354., 356., 358.], dtype=float32) - month(month)int641 2 3 4 5 6 7 8 9 10 11 12
array([ 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12])
- sst(month, lat, lon)float32-1.8 -1.8 -1.8 -1.8 ... nan nan nan
array([[[-1.8000009, -1.8000009, -1.8000009, ..., -1.8000009, -1.8000009, -1.8000009], [-1.8000009, -1.8000009, -1.8000009, ..., -1.8000009, -1.8000009, -1.8000009], [-1.8000009, -1.8000009, -1.8000009, ..., -1.8000009, -1.8000009, -1.8000009], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[-1.8000009, -1.8000009, -1.8000009, ..., -1.8000009, -1.8000009, -1.8000009], [-1.8000009, -1.8000009, -1.8000009, ..., -1.8000009, -1.8000009, -1.8000009], [-1.8000009, -1.8000009, -1.8000009, ..., -1.8000009, -1.8000009, -1.8000009], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[-1.7995025, -1.7995973, -1.7998415, ..., -1.7997988, -1.7996519, -1.7995045], [-1.7995876, -1.7997634, -1.8000009, ..., -1.8000009, -1.7998358, -1.7996247], [-1.8000009, -1.8000009, -1.8000009, ..., -1.8000009, -1.8000009, -1.8000009], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]], dtype=float32)
So we did what we wanted to do: calculate the climatology at every point in the dataset. Let’s look at the data a bit.
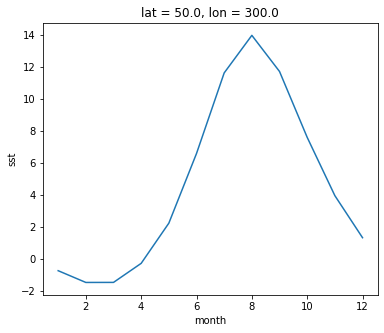
Climatlogy at a specific point in the North Atlantic
ds_mm.sst.sel(lon=300, lat=50).plot()
[<matplotlib.lines.Line2D at 0x7f007c203c40>]
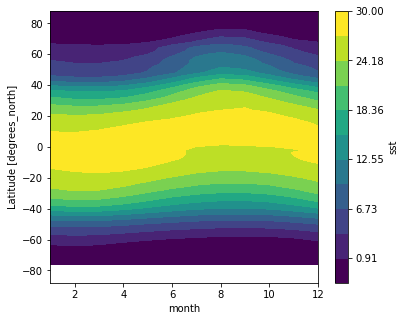
Zonal Mean Climatolgoy
ds_mm.sst.mean(dim='lon').transpose().plot.contourf(levels=12, vmin=-2, vmax=30)
<matplotlib.contour.QuadContourSet at 0x7f007c1e86d0>
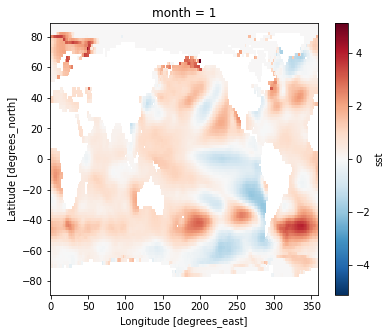
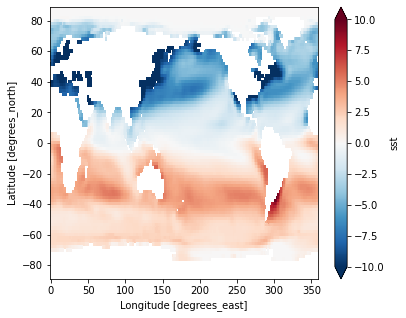
Difference between January and July Climatology
(ds_mm.sst.sel(month=1) - ds_mm.sst.sel(month=7)).plot(vmax=10)
<matplotlib.collections.QuadMesh at 0x7f007c135430>
Transformations¶
Now we want to remove this climatology from the dataset, to examine the residual, called the anomaly, which is the interesting part from a climate perspective. Removing the seasonal climatology is a perfect example of a transformation: it operates over a group, but doesn’t change the size of the dataset. Here is one way to code it
def remove_time_mean(x):
return x - x.mean(dim='time')
ds_anom = ds.groupby('time.month').apply(remove_time_mean)
ds_anom
<xarray.Dataset>
Dimensions: (lat: 89, lon: 180, time: 708)
Coordinates:
* lat (lat) float32 88.0 86.0 84.0 82.0 80.0 ... -82.0 -84.0 -86.0 -88.0
* lon (lon) float32 0.0 2.0 4.0 6.0 8.0 ... 350.0 352.0 354.0 356.0 358.0
* time (time) datetime64[ns] 1960-01-01 1960-02-01 ... 2018-12-01
Data variables:
sst (time, lat, lon) float32 9.537e-07 9.537e-07 9.537e-07 ... nan nan- lat: 89
- lon: 180
- time: 708
- lat(lat)float3288.0 86.0 84.0 ... -86.0 -88.0
- units :
- degrees_north
- long_name :
- Latitude
- actual_range :
- [ 88. -88.]
- standard_name :
- latitude
- axis :
- Y
- coordinate_defines :
- center
array([ 88., 86., 84., 82., 80., 78., 76., 74., 72., 70., 68., 66., 64., 62., 60., 58., 56., 54., 52., 50., 48., 46., 44., 42., 40., 38., 36., 34., 32., 30., 28., 26., 24., 22., 20., 18., 16., 14., 12., 10., 8., 6., 4., 2., 0., -2., -4., -6., -8., -10., -12., -14., -16., -18., -20., -22., -24., -26., -28., -30., -32., -34., -36., -38., -40., -42., -44., -46., -48., -50., -52., -54., -56., -58., -60., -62., -64., -66., -68., -70., -72., -74., -76., -78., -80., -82., -84., -86., -88.], dtype=float32) - lon(lon)float320.0 2.0 4.0 ... 354.0 356.0 358.0
- units :
- degrees_east
- long_name :
- Longitude
- actual_range :
- [ 0. 358.]
- standard_name :
- longitude
- axis :
- X
- coordinate_defines :
- center
array([ 0., 2., 4., 6., 8., 10., 12., 14., 16., 18., 20., 22., 24., 26., 28., 30., 32., 34., 36., 38., 40., 42., 44., 46., 48., 50., 52., 54., 56., 58., 60., 62., 64., 66., 68., 70., 72., 74., 76., 78., 80., 82., 84., 86., 88., 90., 92., 94., 96., 98., 100., 102., 104., 106., 108., 110., 112., 114., 116., 118., 120., 122., 124., 126., 128., 130., 132., 134., 136., 138., 140., 142., 144., 146., 148., 150., 152., 154., 156., 158., 160., 162., 164., 166., 168., 170., 172., 174., 176., 178., 180., 182., 184., 186., 188., 190., 192., 194., 196., 198., 200., 202., 204., 206., 208., 210., 212., 214., 216., 218., 220., 222., 224., 226., 228., 230., 232., 234., 236., 238., 240., 242., 244., 246., 248., 250., 252., 254., 256., 258., 260., 262., 264., 266., 268., 270., 272., 274., 276., 278., 280., 282., 284., 286., 288., 290., 292., 294., 296., 298., 300., 302., 304., 306., 308., 310., 312., 314., 316., 318., 320., 322., 324., 326., 328., 330., 332., 334., 336., 338., 340., 342., 344., 346., 348., 350., 352., 354., 356., 358.], dtype=float32) - time(time)datetime64[ns]1960-01-01 ... 2018-12-01
- long_name :
- Time
- delta_t :
- 0000-01-00 00:00:00
- avg_period :
- 0000-01-00 00:00:00
- prev_avg_period :
- 0000-00-07 00:00:00
- standard_name :
- time
- axis :
- T
- actual_range :
- [19723. 80750.]
- _ChunkSizes :
- 1
array(['1960-01-01T00:00:00.000000000', '1960-02-01T00:00:00.000000000', '1960-03-01T00:00:00.000000000', ..., '2018-10-01T00:00:00.000000000', '2018-11-01T00:00:00.000000000', '2018-12-01T00:00:00.000000000'], dtype='datetime64[ns]')
- sst(time, lat, lon)float329.537e-07 9.537e-07 ... nan nan
array([[[ 9.5367432e-07, 9.5367432e-07, 9.5367432e-07, ..., 9.5367432e-07, 9.5367432e-07, 9.5367432e-07], [ 9.5367432e-07, 9.5367432e-07, 9.5367432e-07, ..., 9.5367432e-07, 9.5367432e-07, 9.5367432e-07], [ 9.5367432e-07, 9.5367432e-07, 9.5367432e-07, ..., 9.5367432e-07, 9.5367432e-07, 9.5367432e-07], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ 9.5367432e-07, 9.5367432e-07, 9.5367432e-07, ..., 9.5367432e-07, 9.5367432e-07, 9.5367432e-07], [ 9.5367432e-07, 9.5367432e-07, 9.5367432e-07, ..., 9.5367432e-07, 9.5367432e-07, 9.5367432e-07], [ 9.5367432e-07, 9.5367432e-07, 9.5367432e-07, ..., 9.5367432e-07, 9.5367432e-07, 9.5367432e-07], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[-4.9746037e-04, -4.0268898e-04, -1.5842915e-04, ..., -2.0110607e-04, -3.4809113e-04, -4.9543381e-04], [-4.1234493e-04, -2.3651123e-04, 9.5367432e-07, ..., 9.5367432e-07, -1.6415119e-04, -3.7527084e-04], [ 9.5367432e-07, 9.5367432e-07, 9.5367432e-07, ..., 9.5367432e-07, 9.5367432e-07, 9.5367432e-07], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]], dtype=float32)
Xarray makes these sorts of transformations easy by supporting groupby arithmetic. This concept is easiest explained with an example:
gb = ds.groupby('time.month')
ds_anom = gb - gb.mean(dim='time')
ds_anom
<xarray.Dataset>
Dimensions: (lat: 89, lon: 180, time: 708)
Coordinates:
* lat (lat) float32 88.0 86.0 84.0 82.0 80.0 ... -82.0 -84.0 -86.0 -88.0
* lon (lon) float32 0.0 2.0 4.0 6.0 8.0 ... 350.0 352.0 354.0 356.0 358.0
* time (time) datetime64[ns] 1960-01-01 1960-02-01 ... 2018-12-01
month (time) int64 1 2 3 4 5 6 7 8 9 10 11 ... 2 3 4 5 6 7 8 9 10 11 12
Data variables:
sst (time, lat, lon) float32 9.537e-07 9.537e-07 9.537e-07 ... nan nan- lat: 89
- lon: 180
- time: 708
- lat(lat)float3288.0 86.0 84.0 ... -86.0 -88.0
- units :
- degrees_north
- long_name :
- Latitude
- actual_range :
- [ 88. -88.]
- standard_name :
- latitude
- axis :
- Y
- coordinate_defines :
- center
array([ 88., 86., 84., 82., 80., 78., 76., 74., 72., 70., 68., 66., 64., 62., 60., 58., 56., 54., 52., 50., 48., 46., 44., 42., 40., 38., 36., 34., 32., 30., 28., 26., 24., 22., 20., 18., 16., 14., 12., 10., 8., 6., 4., 2., 0., -2., -4., -6., -8., -10., -12., -14., -16., -18., -20., -22., -24., -26., -28., -30., -32., -34., -36., -38., -40., -42., -44., -46., -48., -50., -52., -54., -56., -58., -60., -62., -64., -66., -68., -70., -72., -74., -76., -78., -80., -82., -84., -86., -88.], dtype=float32) - lon(lon)float320.0 2.0 4.0 ... 354.0 356.0 358.0
- units :
- degrees_east
- long_name :
- Longitude
- actual_range :
- [ 0. 358.]
- standard_name :
- longitude
- axis :
- X
- coordinate_defines :
- center
array([ 0., 2., 4., 6., 8., 10., 12., 14., 16., 18., 20., 22., 24., 26., 28., 30., 32., 34., 36., 38., 40., 42., 44., 46., 48., 50., 52., 54., 56., 58., 60., 62., 64., 66., 68., 70., 72., 74., 76., 78., 80., 82., 84., 86., 88., 90., 92., 94., 96., 98., 100., 102., 104., 106., 108., 110., 112., 114., 116., 118., 120., 122., 124., 126., 128., 130., 132., 134., 136., 138., 140., 142., 144., 146., 148., 150., 152., 154., 156., 158., 160., 162., 164., 166., 168., 170., 172., 174., 176., 178., 180., 182., 184., 186., 188., 190., 192., 194., 196., 198., 200., 202., 204., 206., 208., 210., 212., 214., 216., 218., 220., 222., 224., 226., 228., 230., 232., 234., 236., 238., 240., 242., 244., 246., 248., 250., 252., 254., 256., 258., 260., 262., 264., 266., 268., 270., 272., 274., 276., 278., 280., 282., 284., 286., 288., 290., 292., 294., 296., 298., 300., 302., 304., 306., 308., 310., 312., 314., 316., 318., 320., 322., 324., 326., 328., 330., 332., 334., 336., 338., 340., 342., 344., 346., 348., 350., 352., 354., 356., 358.], dtype=float32) - time(time)datetime64[ns]1960-01-01 ... 2018-12-01
- long_name :
- Time
- delta_t :
- 0000-01-00 00:00:00
- avg_period :
- 0000-01-00 00:00:00
- prev_avg_period :
- 0000-00-07 00:00:00
- standard_name :
- time
- axis :
- T
- actual_range :
- [19723. 80750.]
- _ChunkSizes :
- 1
array(['1960-01-01T00:00:00.000000000', '1960-02-01T00:00:00.000000000', '1960-03-01T00:00:00.000000000', ..., '2018-10-01T00:00:00.000000000', '2018-11-01T00:00:00.000000000', '2018-12-01T00:00:00.000000000'], dtype='datetime64[ns]') - month(time)int641 2 3 4 5 6 7 ... 6 7 8 9 10 11 12
array([ 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, ... 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12])
- sst(time, lat, lon)float329.537e-07 9.537e-07 ... nan nan
array([[[ 9.5367432e-07, 9.5367432e-07, 9.5367432e-07, ..., 9.5367432e-07, 9.5367432e-07, 9.5367432e-07], [ 9.5367432e-07, 9.5367432e-07, 9.5367432e-07, ..., 9.5367432e-07, 9.5367432e-07, 9.5367432e-07], [ 9.5367432e-07, 9.5367432e-07, 9.5367432e-07, ..., 9.5367432e-07, 9.5367432e-07, 9.5367432e-07], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[ 9.5367432e-07, 9.5367432e-07, 9.5367432e-07, ..., 9.5367432e-07, 9.5367432e-07, 9.5367432e-07], [ 9.5367432e-07, 9.5367432e-07, 9.5367432e-07, ..., 9.5367432e-07, 9.5367432e-07, 9.5367432e-07], [ 9.5367432e-07, 9.5367432e-07, 9.5367432e-07, ..., 9.5367432e-07, 9.5367432e-07, 9.5367432e-07], ... [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]], [[-4.9746037e-04, -4.0268898e-04, -1.5842915e-04, ..., -2.0110607e-04, -3.4809113e-04, -4.9543381e-04], [-4.1234493e-04, -2.3651123e-04, 9.5367432e-07, ..., 9.5367432e-07, -1.6415119e-04, -3.7527084e-04], [ 9.5367432e-07, 9.5367432e-07, 9.5367432e-07, ..., 9.5367432e-07, 9.5367432e-07, 9.5367432e-07], ..., [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan], [ nan, nan, nan, ..., nan, nan, nan]]], dtype=float32)
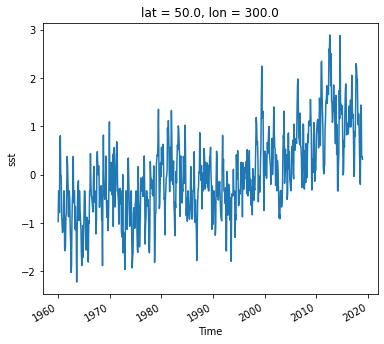
Now we can view the climate signal without the overwhelming influence of the seasonal cycle.
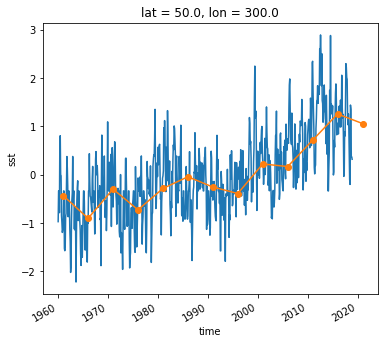
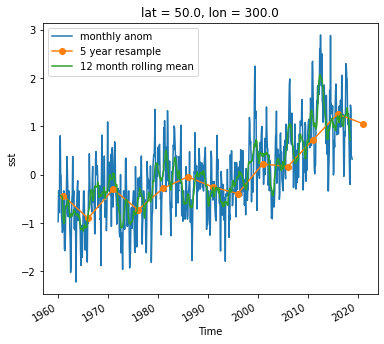
Timeseries at a single point in the North Atlantic
ds_anom.sst.sel(lon=300, lat=50).plot()
[<matplotlib.lines.Line2D at 0x7f007c2579d0>]
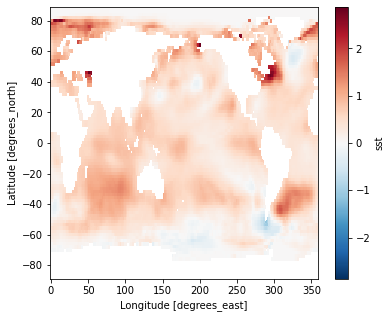
Difference between Jan. 1 2018 and Jan. 1 1960
(ds_anom.sel(time='2018-01-01') - ds_anom.sel(time='1960-01-01')).sst.plot()
<matplotlib.collections.QuadMesh at 0x7f00777f76a0>